AlphaFold helps Gemma Atkinson discover bacterial immune systems
Gemma Atkinson uses all kinds of NAISS resources to find new bacterial immune systems. She says researchers in Sweden are lucky to have large-scale computing and storage resources available for free, and that this fact also helps her recruit new colleagues.
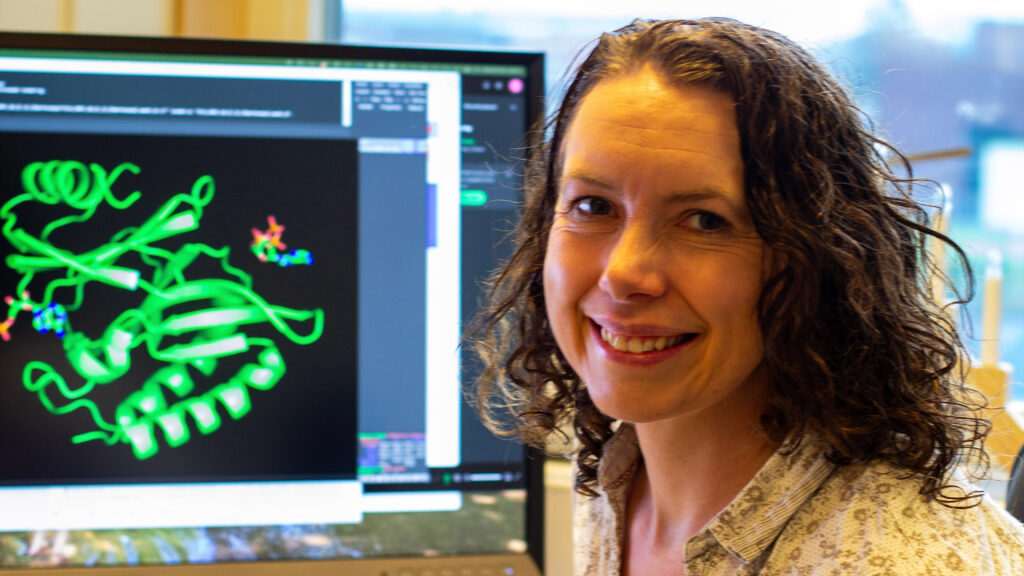
Gemma Atkinson is a senior lecturer at Lund University and leader of a bioinformatics team at the Atkinson Lab. In cooperation with the experimental lab of her colleague, senior lecturer Vasili Hauryliuk, she is searching through bacterial genomes to find new immune systems.
Studying these immune systems is important for understanding how viruses shape our environments and microbiomes, and how immune systems are evolving.
“The viruses which infect bacteria, so called bacteriophages, or phages, can kill pathogenic bacteria that are resistant to antibiotics. Phage therapy is seen as a promising way out of the current antibiotic resistance crisis, both in the clinic and in agriculture,” Gemma Atkinson explains.
New infrastructure to assist other researchers
Bacterial immune systems are usually made of proteins, which fold into specific 3D shapes called structures. Figuring out these structures used to be a huge challenge that could take years and require expensive lab equipment. But thanks to AlphaFold, the AI-powered protein prediction programme developed by DeepMind, and other similar AI models, excellent structures can now be produced for many proteins in just a fraction of the time.
Gemma Atkinson says it completely changed the way she works. AlphaFold has been so successful for her group that she became inspired to help other researchers. She set up a national infrastructure, LU-Fold, to assist them.
“It’s hard to understate just how important this breakthrough is. The Nobel Prize in chemistry is really well deserved.”
However, running these models are still computationally challenging, and she happily admits that she is a heavy user of NAISS resources.
“We need a lot of power, so we use everything. We are doing so much! For AlphaFold, it is almost entirely AI/GPU resources. For the multiple sequence alignments of proteins that we use to predict the structure, we need CPUs. So we do that on Tetralith usually.”
Free resources a selling point when recruiting
She says she sometimes hits the roof – not just in terms of hours used, but also storage, as these jobs generate a lot of files. She would also like to see a more streamlined application procedure.
“But the fact that these amazing computational resources are available for free and we just have to apply for them, I think that we are really lucky in Sweden to have that. This is a big selling point as well. When computational biologists are interested in joining my group I can send them the link to the resources that are available via NAISS, and this quite often helps people make their mind up.”